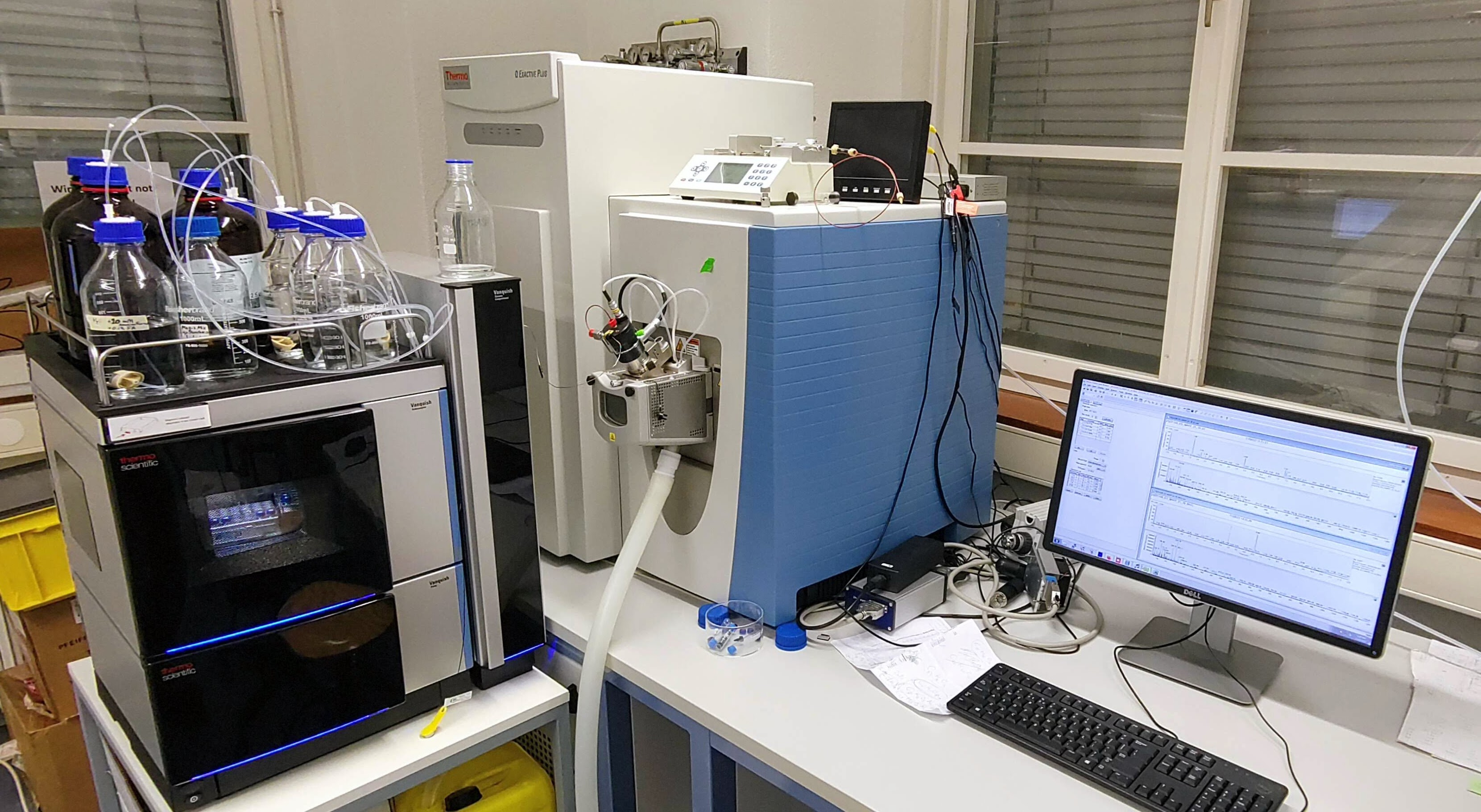
The Q Exactive HF-X instrument (coupled to an Easy-nLC 1200, Thermo Fisher Scientific) is used for state-of-the-art PTM analyses of peptides such as ubiquitinations and phosphorylations.
Metabolomics and Proteomics Platform (MAPP)
The Metabolomics and Proteomics Platform (MAPP) is a core service of the Department of Biology at the University of Fribourg launched in 2017. The proteomics unit is embedded within the groups of Prof. Jörn Dengjel (expert in proteomics) and Dr Dieter Kressler (expert in protein-protein interactions). The metabolomics unit is associated to the group of Dr Pierre-Marie Allard (expert in metabolomics and natural products chemistry).
We provide our expertise, instrumentation, and manpower to enable a state-of-the-art implementation of your metabolomic and proteomic analyses. We support you in the planning and execution of your experiments, including the establishment and development of methods, sample preparation, data acquisition, and data analysis. Moreover, we also provide the tools for conducting downstream protein-protein interaction analyses (e.g. yeast two-hybrid and in vitro binding assays).
If you have any question regarding proteome or metabolome analysis, do not hesitate to contact with us via email or directly by coming to our offices or labs.
We encourage you to contact us before you start your experiment.
Our platform currently encompasses:
- Two nanoLC-ESI-MS/MS systems (Q Exactive Plus and Q Exactive HF-X, both Thermo Fisher Scientific)
- Off-line Dionex Ultimate 3000 UHPLC system
- Gas Chromatograph-Quadrupole-Time-of-Flight (Agilent Technologies)
- Gas Chromatograph-Mass Spectrometer (Agilent Technologies)
- Gas Chromatograph-Flame Ionization Detector (Agilent Technologies)
- High-Performance Liquid Chromatography/Diode Array Detector/Fluorescence Detector (Agilent Technologies)
- High-Performance Liquid Chromatography/Fraction Collector (Agilent Technologies)
-
Equipment
Omics
The Q Exactive Plus instrument (coupled to an Easy-nLC 1000, Thermo Fisher Scientific) is used for state-of-the-art proteomics and metabolomics experiments.
Standard workflows include: Data are recorded with the Xcalibur software (Thermo Scientific), processed with MaxQuant software package (MPI of Biochemistry, Martinsried), using its integrated Andromeda search engine for peptide identification. Statistical analyses are performed with the Perseus software (MPI of Biochemistry, Martinsried). Metabolomics data are analysed using the MZmine software with online access to several metabolite databases.
Metabolite Profiling
In addition, for metabolite profiling purposes, compound purification, and purity checks HPLC with DAD and FLD as well as GC-FID or GC-EI-MS (all Agilent Technologies) will be used.
-
Proteomics
SERVICES AND PRICING
Please contact us before you start your experiments for discussing the design as well as for additional services (e.g. quantitative proteomics by SILAC labeling).
Service University of Fribourg External academics Private (commercial) Single run (sample is ready for injection on LC-MS) 50.- 80.- 200.- Single sample analysis (incl. sample preparation) per LC-MS run* 60.- 100.- 240.- Phosphoproteomics (incl. enrichment and LC-MS run) per sample 100.- 120.- 280.- Direct infusion (w/o LC) e.g. for intact mass determination 80.- 100.- 240.- * a protein extract will be divided into 3-10 different fractions (depending on complexity) that are analysed separately, e.g. affinity pulldown (medium complexity) will end up in 5 LC-MS runs - in total 400.- CHF.
DOCUMENTS
-
Metabolomics
SERVICES
UHPLC-HRMS (QE Plus)
• Untargeted metabolomics (DDA/DIA)
• Targeted metabolomics (e.g. plant hormones)
• Identification of compounds by mass spectrometry
• Compound purity checkGC-qTOF (Agilent 7200)
Full-spectrum, high-resolution, accurate-mass data for profiling, identifying and quantifying volatile compounds (+/- derivatization)
• High-resolution mass identification of analytes
• Metabolite profiling (e.g. bacterial volatilome, essential oils, ...)
• Metabolite quantificationGC-FID/GC-MS (Agilent 7890)
Profiling, identifying and quantifying volatile compounds (+/- derivatization)
• Fatty acid Methyl Ester
• Steroids
• CannabinoidsHPLC-DAD/FLD (Agilent 1260)
Equipped for both normal (silica, diol) and reverse-phase (C18) HPLC
- Quantification of hydrophylic/hydrophobic compounds with light absorbing/fluorescent properties
- Compound purity
- Tocochromanol (vitamin E), carotenoid (pro-vitamin A)
- Camalexin
- Salicylic acid
- Neurotransmitters
Fraction collector (Agilent Infinity II)
Automated fraction collector connected to the Agilent 1260 HPLC system
• HPLC (normal/reverse phase) purification of compound(s) of interest
• Peak collection triggered by time/acquisition signal(s)
• Automated (sequence-based) purification
In addition to these compounds that we routinely analyze, we can develop new protocols for your research. We also offer our expertise regarding mass spectrometric data organization and interpretation in particular regarding metabolite annotation. We are happy to discuss your specific needs and adapt to your research project. For metabolomics related projects please contact Dr Pierre-Marie Allard directly.
-
Training and Education
If you are interested, we are happy to show you how the samples are processed and analysed, which will enable you to perform the complete sample processing and data analysis. This makes especially sense if you intend to carry out metabolomic or proteomic analyses on a regular basis. However, the instrumentation is only operated by us.
In addition, team members give also lectures and practical courses covering distinct topics of metabolomic and proteomic analyses.
Jörn Dengjel
Prof. and Group Leader
PER 05 - 0.312
+41 26 300 8631
Dieter Kressler
PhD, group leader, head of proteomics platform
PER 05 - 0.316d
+41 26 300 8645
Pierre-Marie Allard
PER 04 - 0.104
+41 26 300 8808
Michael Stumpe
PER 05 - 0.317b
+41 26 300 8640