Daniele Silvestro
Assistant Professor
PER 05 - 0.349A
+41 26 300 8857
E-mail
We develop computational methods and software to study evolutionary processes across different organisms including plants, vertebrates, and invertebrate groups.
We use phylogenetic and fossil data to answer different questions in macroevolution:
The main statistical frameworks we use to tackle these questions are Bayesian methods and machine learning. We develop and implement new models in open source software.
We also develop new methods and software to predict extinction risk in modern species, to model the dynamics of the ongoing biodiversity crisis, and to optimize conservation efforts using artificial intelligence.
The group is funded by the Eccellenza program from the Swiss National Science Foundation (PCEFP3_187012).
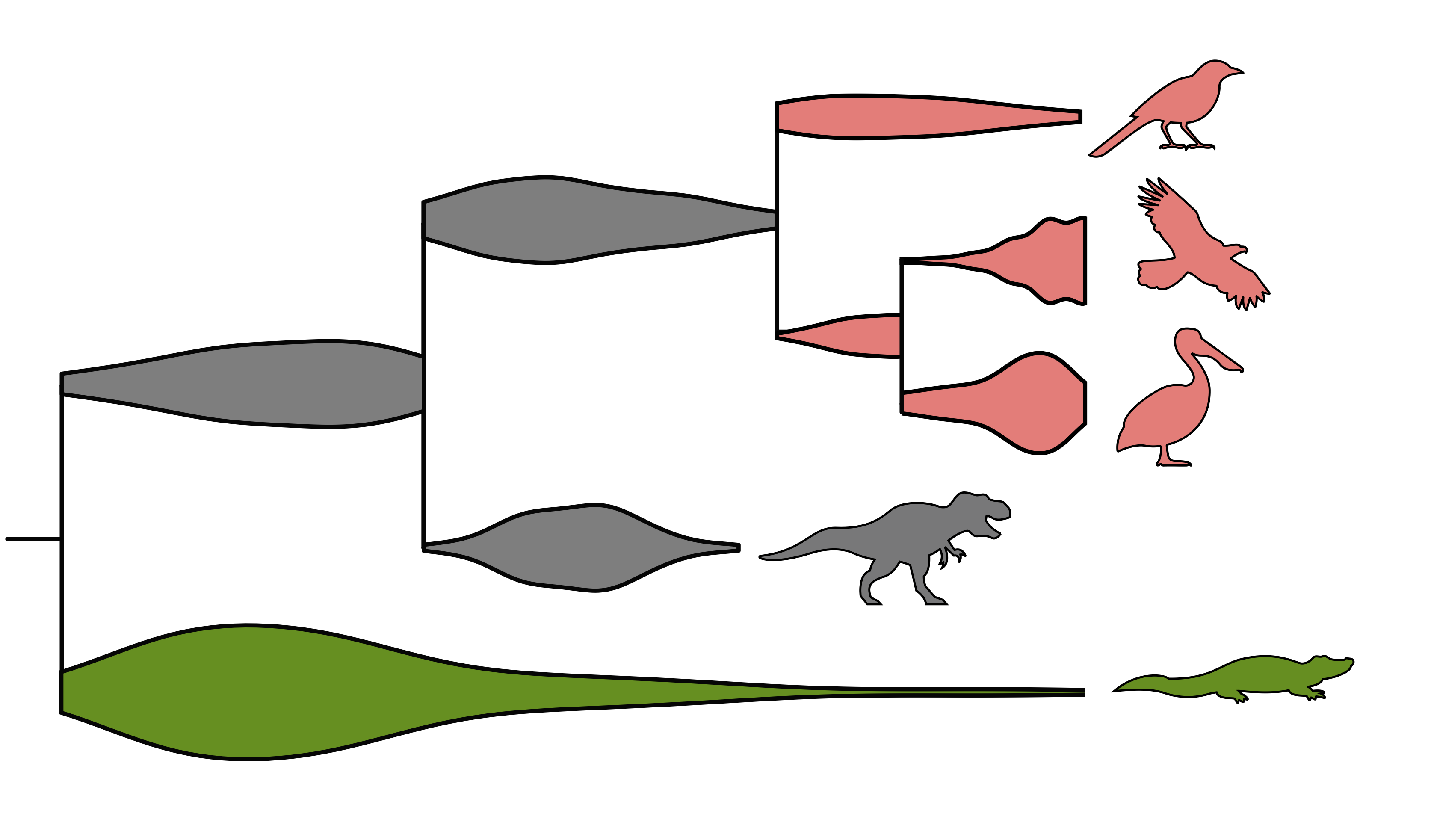
We are developing new methods to analyze fossil occurrence data linking intraspecific population dynamics with speciation and extinction models. Establishing these links will allow us to better understand the causes behind the rise and fall of species and to bridge over the long-standing divide between micro- and macro- evolutionary models. We use data from different plant and animal clades to tackle the following questions:
Answering these questions requires new models explicitly incorporating population dynamics, speciation and extinction as well as preservation and sampling processes.
One of the most striking patterns in hyper-diverse forests such as Amazonia, is the imbalance between few hyper-dominant plant species making up the majority of all trees and the thousands of very rare species with few individuals often scattered across large regions1. The evolutionary history that lead to this imbalance remains a mystery.
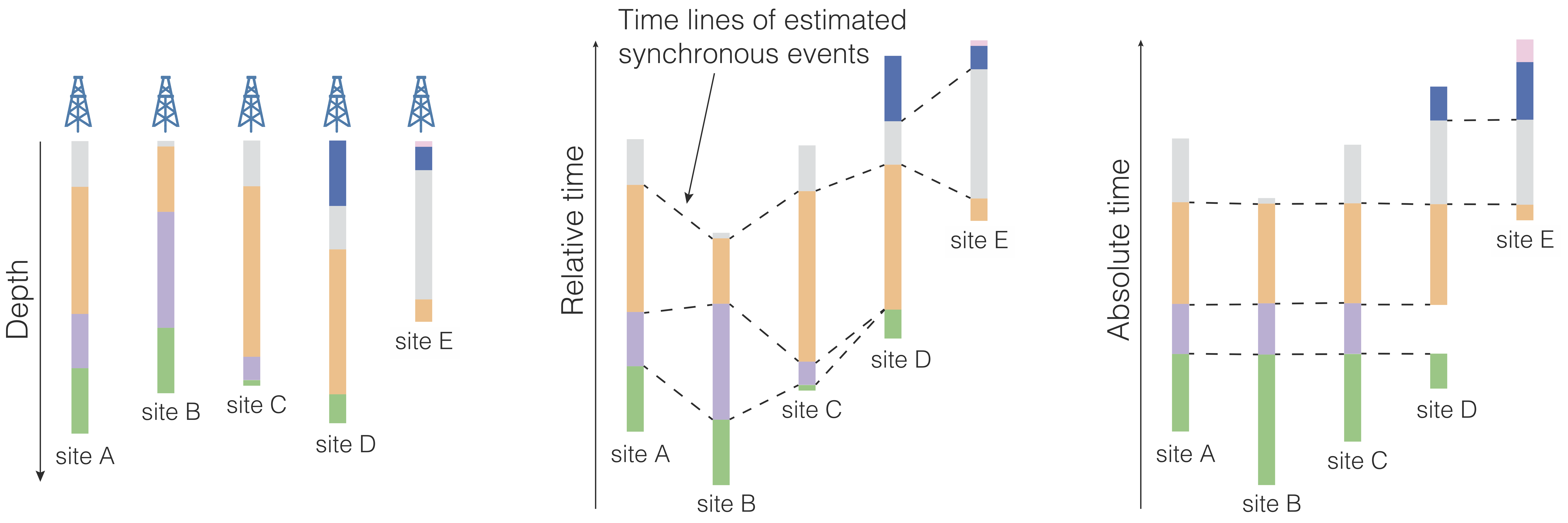
The fossil pollen record provides the most abundant and detailed account of past vegetation and its changes through time, with hundreds of recognized morpho-species and millions of occurrences. Pollen data can be used to track ecosystem changes (e.g. from tropical rain forest to savanna), the origination and extinction of plant lineages and their changes in relative abundance. Yet the analysis of this wealth of data presents two major challenges: the identification and digitization of large numbers of pollen grains and the presence of sampling and preservation biases that may hamper a direct reading of the fossil record.
Within this project we are developing new identification methods for pollen using deep learning and neural networks. We are also implementing new Bayesian methods integrating biostratigraphy, population dynamics models, and diversification and extinction processes
We apply these methods to the rich fossil pollen data of South America, which encompasses millions of records and spans the past 140 million years and aim to understand the origin and evolution of tropical forests.
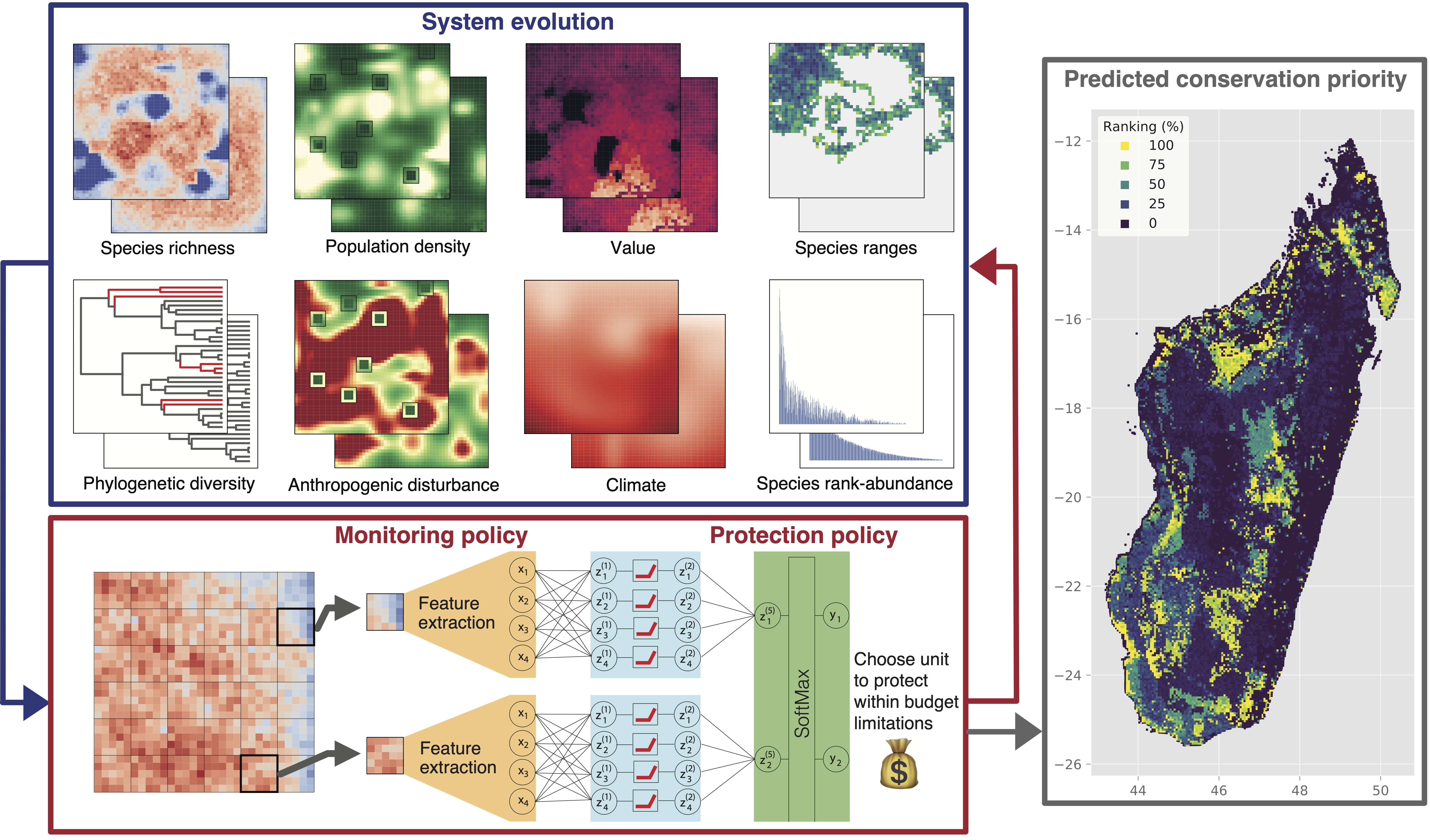
More then a million species might be facing extinction in the near future due to increasing anthropogenic pressure on the natural world and climate change. We develop AI tools to assess species extinction risk, model future biodiversity loss, and optimize systematic conservation planning.
Assistant Professor
PER 05 - 0.349A
+41 26 300 8857
E-mail
Department of Biology
Chemin du Musée 10
CH-1700 Fribourg
Switzerland